Table 1 Statistics of domain networks The domain networks of S. cerevisiae, C. elegans, D. melanogaster, M. musculus and H. sapiens all contain a major component (mc) which incorporates the majority of the domains ( ), coexisting with many small, connected components (N
cc
). The number of edges (
), coexisting with many small, connected components (N
cc
). The number of edges (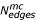 ) of the main component, the mean node degree ⟨k⟩ and the mean clustering coefficient ⟨C⟩ gradually increase with the organisms level of development. Referring to Fig. 1, the frequency distributions of the number N of domains per protein (Fig. 1b) follows a power-law P(N) ~ N-δ. Similarly, we find a power-law in the distributions of the occurrence of domains N and their mean degree ⟨k⟩ in the organism specific co-occurrence networks k ~ Nε(Fig. 1c). The degree distributions of the organisms specific co-occurrence networks can be approximated by a power-law P(k) ~ k-θ(Fig. 1d). Similarly, we approximated the degree dependence of the clustering coefficient by a generalized Zipf-law ⟨C(k)⟩ = α(β + k)-γ(Fig. 1e).
) of the main component, the mean node degree ⟨k⟩ and the mean clustering coefficient ⟨C⟩ gradually increase with the organisms level of development. Referring to Fig. 1, the frequency distributions of the number N of domains per protein (Fig. 1b) follows a power-law P(N) ~ N-δ. Similarly, we find a power-law in the distributions of the occurrence of domains N and their mean degree ⟨k⟩ in the organism specific co-occurrence networks k ~ Nε(Fig. 1c). The degree distributions of the organisms specific co-occurrence networks can be approximated by a power-law P(k) ~ k-θ(Fig. 1d). Similarly, we approximated the degree dependence of the clustering coefficient by a generalized Zipf-law ⟨C(k)⟩ = α(β + k)-γ(Fig. 1e).
organism | N cc |
|
| ⟨k⟩ | ⟨C⟩ | δ | ε | θ | α | β | γ |
|---|---|---|---|---|---|---|---|---|---|---|---|
H. sapiens | 172 | 733 | 4,048 | 5.52 | 0.42 | 2.1 | 0.5 | 1.4 | 61.6 | 17.9 | 1.5 |
M. musculus | 173 | 668 | 3,566 | 5.34 | 0.43 | 2.5 | 0.5 | 1.5 | 67.5 | 17.1 | 1.5 |
D. melanogaster | 172 | 506 | 2,274 | 4.49 | 0.39 | 2.3 | 0.4 | 1.6 | 1,407.8 | 34.7 | 2.7 |
C. elegans | 167 | 495 | 2,120 | 4.28 | 0.34 | 2.4 | 0.4 | 1.7 | 3,886.7 | 30.9 | 2.5 |
S. cerevisiae | 177 | 175 | 516 | 2.95 | 0.32 | 2.6 | 0.3 | 2.0 | 39.4 | 11.3 | 1.5 |
 ), coexisting with many small, connected components (N
cc
). The number of edges (
), coexisting with many small, connected components (N
cc
). The number of edges (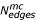 ) of the main component, the mean node degree
) of the main component, the mean node degree